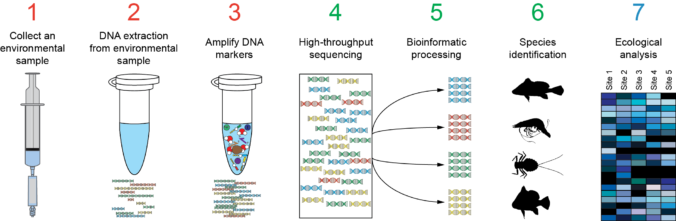
In the previous post I explained the fundamentals about the Amplicon Sequencing (AS) technique, today I will show some current and future applications in Metagenomics and Metabarcoding.
Metagenomics (also referred to as ‘environmental’ or ‘community’ genomics) is the study of genetic material recovered directly from environmental samples. This discipline applies a suite of genomic technologies and bioinformatics tools to directly access the genetic content of entire communities of organisms. Usually we use the term metabarcoding when we apply the amplicon sequencing approach in metagenomics studies, also metagenomics term if preferred when we study full genomes, not only few gene regions.
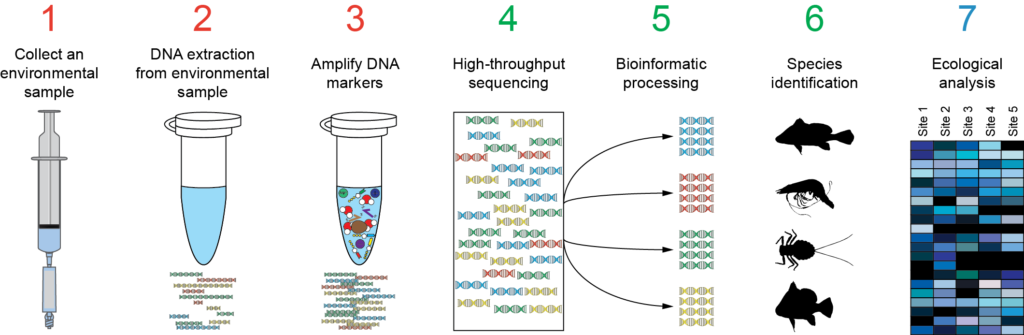
Metabarcoding workflow. Source: http://www.naturemetrics.co.uk
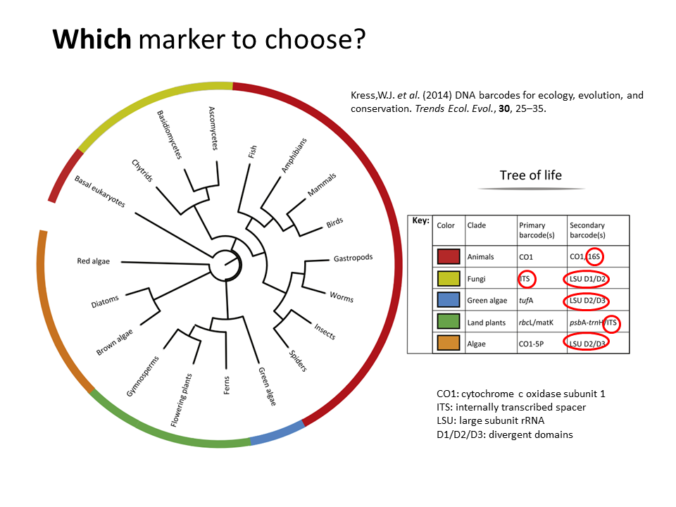
For metabarcoding, 16S rRNA gene is the most common universal DNA barcode (marker) used to identify with great accuracy species from across the Tree of Life, but other genes as: cytochrome c oxidase subunit 1 (CO1), rRNA (16S/18S/28S), plant specific ones (rbcL, matK, and trnH-psbA) and gene regions as: internal transcribed spacers (ITSs) (Kress et al. 2014; Joly et al. 2014). The previous genes have mutation rates fast enough to discriminate close species and at the same time they are stable enough to identify individuals of the same specie.
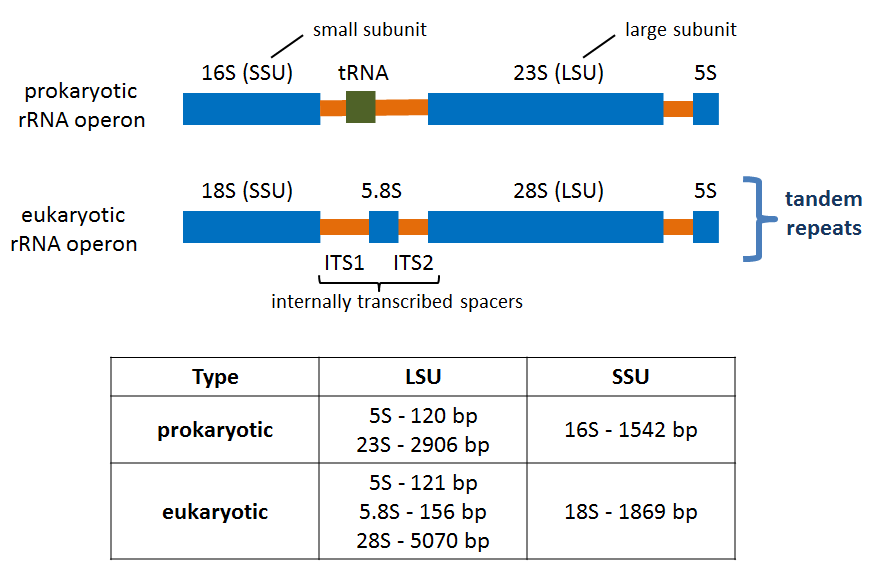
Prokaryotic and eukaryotic rRNA operons
A perfect metagenomics barcode/marker should…
- be present in all the organisms, in all the cells
- have variable sequence among different species
- be conserved among individuals of the same species
- be easy to amplify and not too long for sequencing
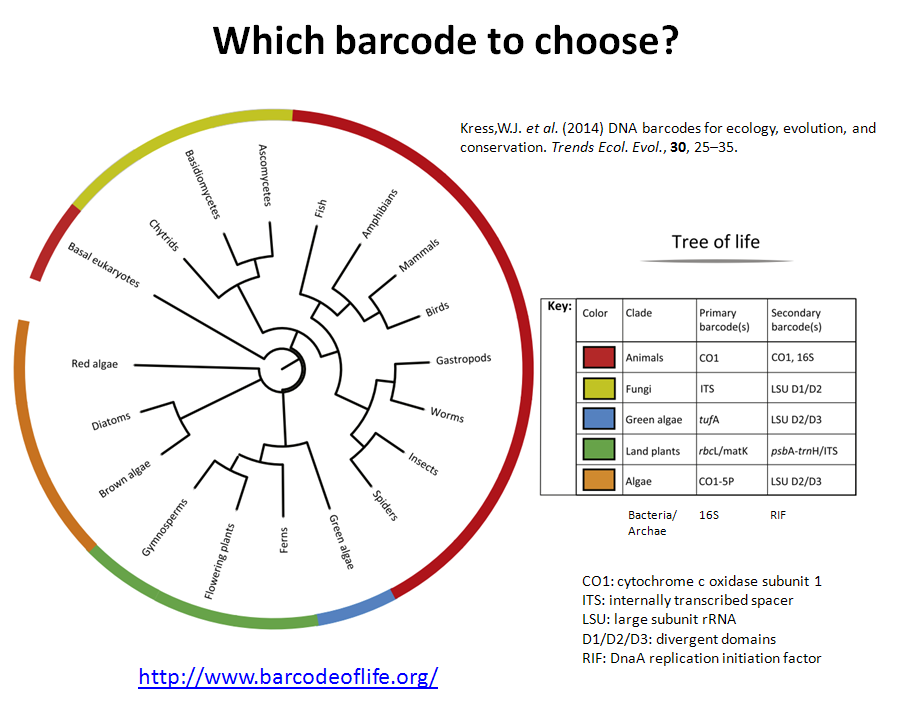
Recommended DNA barcodes for metagenomics
The pioneer metabarcoding study of Sogin et al. 2006 to decipher the microbial diversity in the deep sea used as barcode the V6 hypervariable region of the rRNA gene. Sogin et al. sequenced around 118,000 PCR amplicons from environmental DNA preparations and unveiled thousand of new species not known before.
Observe that in metabarcoding we cannot use DNA tags to pick out single individuals, but to identify different samples (of water, soil, air…).